It is critical to understand details of the global carbon and nitrogen cycles given that they control atmospheric concentrations of the three major greenhouse gases: CO2, CH4 and N2O.
Many of the metalloenzymes that process such gaseous small molecules employ active sites comprised of metal clusters that mediate multi-electron/multi-proton redox catalysis. Our group’s contributions are to build and study synthetic model compounds that mimic the structure, spectroscopy, and reactivity of the metal cluster active sites.
Reactivity studies of the synthetic models map structure-function relationships governing their inorganic functional groups and elucidate key second coordination sphere effects that can be mimicked in synthetic catalyst designs.
Among our targeted active sites have been the O2Mo(µ2-S)Cu cluster in aerobic carbon monoxide dehydrogenase (CODH) and the Cu4(µ4-S) cluster in nitrous oxide reductase (N2OR). Based on these initial studies, we have ongoing projects in the areas of multicopper clusters and molybdo-/tungstoenzyme mimics that emphasize CO2/N2O reduction reactions and C–H activation processes.
Copper clusters for CO2 and N2O reduction reactions.
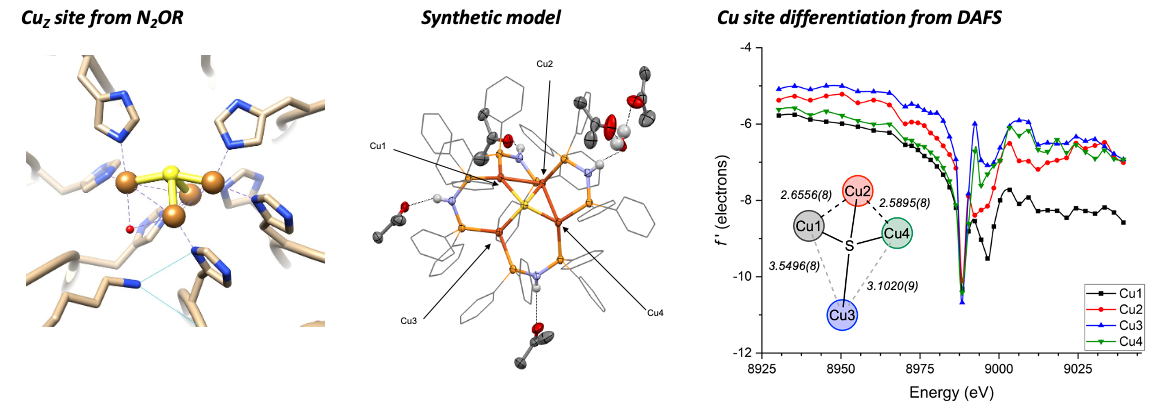
Starting in 2014, our group has been interested in synthesizing mimics of the tetranuclear CuZ catalytic site of nitrous oxide reductase (N2OR).
We have constructed several structurally faithful Cu4(µ4-S) model compounds, including cases that also promote single-turnover, biomimetic N2O reduction
Our published work has used a combination of NMR, EPR, and XANES spectroscopies to analyze the electronic structures of the clusters. To correlate Cu site-specific electronic structure to N2O reactivity behavior, we recently began probing these compounds by resonant X-ray diffraction anomalous fine structure (DAFS) analysis, an advanced crystallography technique that enables X-ray absorption profiles of crystallographically independent metal sites within a cluster to be deconvoluted.
Although DAFS has been used for other metals including analyses of nitrogenase at the Fe K-edge, we are the first group to report DAFS experiments at the Cu K-edge. We have used DAFS to reveal that the reactive 4Cu(I) redox state of a model cluster requires site differentiation (i.e., incipient Cu 𝛿+···Cu𝛿- pairs) induced by structural reorganization in order to activate N2O.
Molybdo- and tungstoenzyme mimics for CO2 and C–H activation reactions.
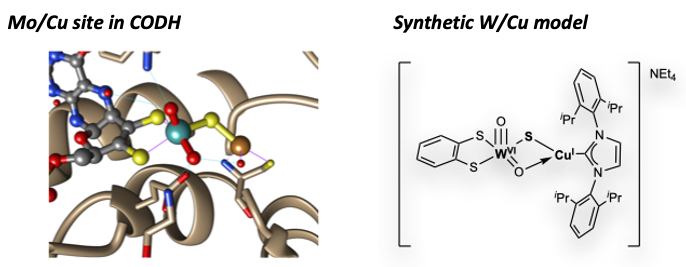
Among the superfamily of oxotransferase enzymes featuring molybdopterin or tungstopterin active sites, the aerobic carbon monoxide dehydrogenase (CODH) enzyme is unique in that its active site features a heterobinuclear structure in which the canonical molybdopterin is closely associated with an unsaturated Cu(I) center via an unsupported Mo–S–Cu bridge.
Given the importance of CODH reactivity, i.e., the 2H+/2e- interconversion of CO and CO2, to the global carbon cycle, it is critical to understand the role of heterobinuclearity in this case.
Our report in 2020 of a model complex with a W–S–Cu bridge is the closest structural mimic discovered to date and represented a landmark after two decades of synthetic efforts by the bioinorganic chemistry community.
The inability of our structural mimic to achieve biomimetic reactivity was ascribed to the lack of second coordination sphere effects required to stabilize the spatially separated M(VI)=O/Cu(I) structure found in the native system. Ongoing synthetic work in our lab, therefore, is targeting second-generation model systems that incorporate Lewis acidic groups and/or hydrogen bond donors in the second coordination sphere.
Additionally, in pursuing this work, we have begun to take advantage of mononuclear Mo and W precursor compounds to build synthetic models of the active sites in mononuclear molybdo- and tungstopterin enzymes.
For example, the aerobic CODH is part of the xanthine oxidase (XO) family of oxotransferases. We are particularly intrigued by XO itself, as its role is to promote chemoselective C–H oxidation of a bespoke substrate.
Similarly, members of the DMSO reductase family of oxotransferases are known to catalyze oxidation of thermodynamically robust C(sp3)–H bonds and represent targets of ongoing synthetic modeling efforts in our labs.
Another member of the DMSO reductase family is the set of formate dehydrogenases (FDHs), which we have begun to model to understand the divergent selectivity of mononuclear FDHs vs. the heterobinuclear CODH.

